Source - http://dienekes.blogspot.fr/
A choice quote:
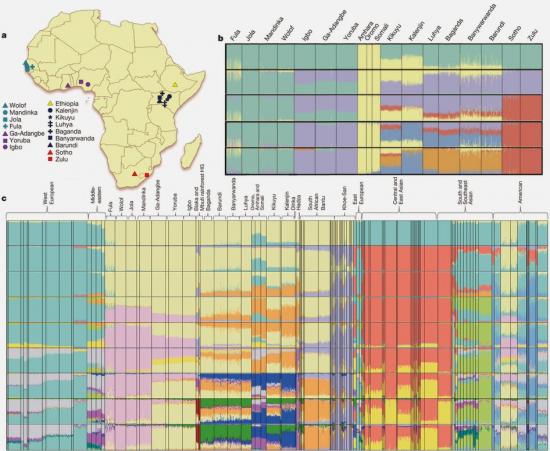
To assess the effect of gene flow on population differentiation in SSA, we masked Eurasian ancestry across the genome (Supplementary Methods and Supplementary Note 6). This markedly reduced population differentiation, as measured by a decline in mean pairwise FST from 0.021 to 0.015 (Supplementary Note 6), suggests that Eurasian ancestry has a substantial impact on differentiation among SSA populations. We speculate that residual differentiation between Ethiopian and other SSA populations after masking Eurasian ancestry (pairwise FST = 0.027) may be a remnant of East African diversity pre-dating the Bantu expansion10.
I think this should be highlighted for a couple of reasons.
1. In too many papers to count, decreasing genetic diversity from East Africa was taken as evidence of an origin ofH. sapiens in that locality and its expansion from there to Eurasia. This "East Africa=cradle of mankind" theory has, as far as I can tell, nothing really to stand on. Granted, the oldest anatomically modern human remains have been found in East Africa 200-150 thousand years ago. But, the fact that old sapiens have been found in East Africa and not elsewhere is easily explained by the excellent conditions for preservation (as opposed, e.g., deserts or rainforests of Africa or elsewhere), and by the extraordinary effort by palaeoanthropologists in that area. One also needs to overlook a century of physical anthropology that concluded that East Africa was a contact zone between Caucasoids and Sub-Saharan Africans. We now know that there is no deep lineage of humans in modern east Africans. Take out the Eurasian ancestry and only a paltry Fst=0.027 remains with other Sub-Saharan Africans, a fraction of the Fst between, say, Europeans and East Asians.
2. There has been enormous literature about phenotypic variation in Africans. The ultra-migrationism of old was replaced by ultra-selectionism that sought to explain every phenotypic marker of Eurasian admixture in Africa not as evidence of such admixture, but as a parallel process of evolution whereby some Africans tended to resemble some Eurasians not because of admixture but because of adaptation to similar environmental conditions.
But:

This suggests that a large proportion of differentiation observed among African populations could be due to Eurasian admixture, rather than adaptation to selective forces (Supplementary Note 6).
This study also confirms the presence of Eurasian admixture in the Yoruba
Our finding of ancient Eurasian admixture corroborates findings of non-zero Neanderthal ancestry in Yoruba, which is likely to have been introduced through Eurasian admixture and back migration, possibly facilitated by greening of the Sahara desert during this period13, 14.
Nature (2014) doi:10.1038/nature13997
The African Genome Variation Project shapes medical genetics in Africa
Deepti Gurdasani, Tommy Carstensen, Fasil Tekola-Ayele, Luca Pagani, Ioanna Tachmazidou, et al.
Given the importance of Africa to studies of human origins and disease susceptibility, detailed characterization of African genetic diversity is needed. The African Genome Variation Project provides a resource with which to design, implement and interpret genomic studies in sub-Saharan Africa and worldwide. The African Genome Variation Project represents dense genotypes from 1,481 individuals and whole-genome sequences from 320 individuals across sub-Saharan Africa. Using this resource, we find novel evidence of complex, regionally distinct hunter-gatherer and Eurasian admixture across sub-Saharan Africa. We identify new loci under selection, including loci related to malaria susceptibility and hypertension. We show that modern imputation panels (sets of reference genotypes from which unobserved or missing genotypes in study sets can be inferred) can identify association signals at highly differentiated loci across populations in sub-Saharan Africa. Using whole-genome sequencing, we demonstrate further improvements in imputation accuracy, strengthening the case for large-scale sequencing efforts of diverse African haplotypes. Finally, we present an efficient genotype array design capturing common genetic variation in Africa.